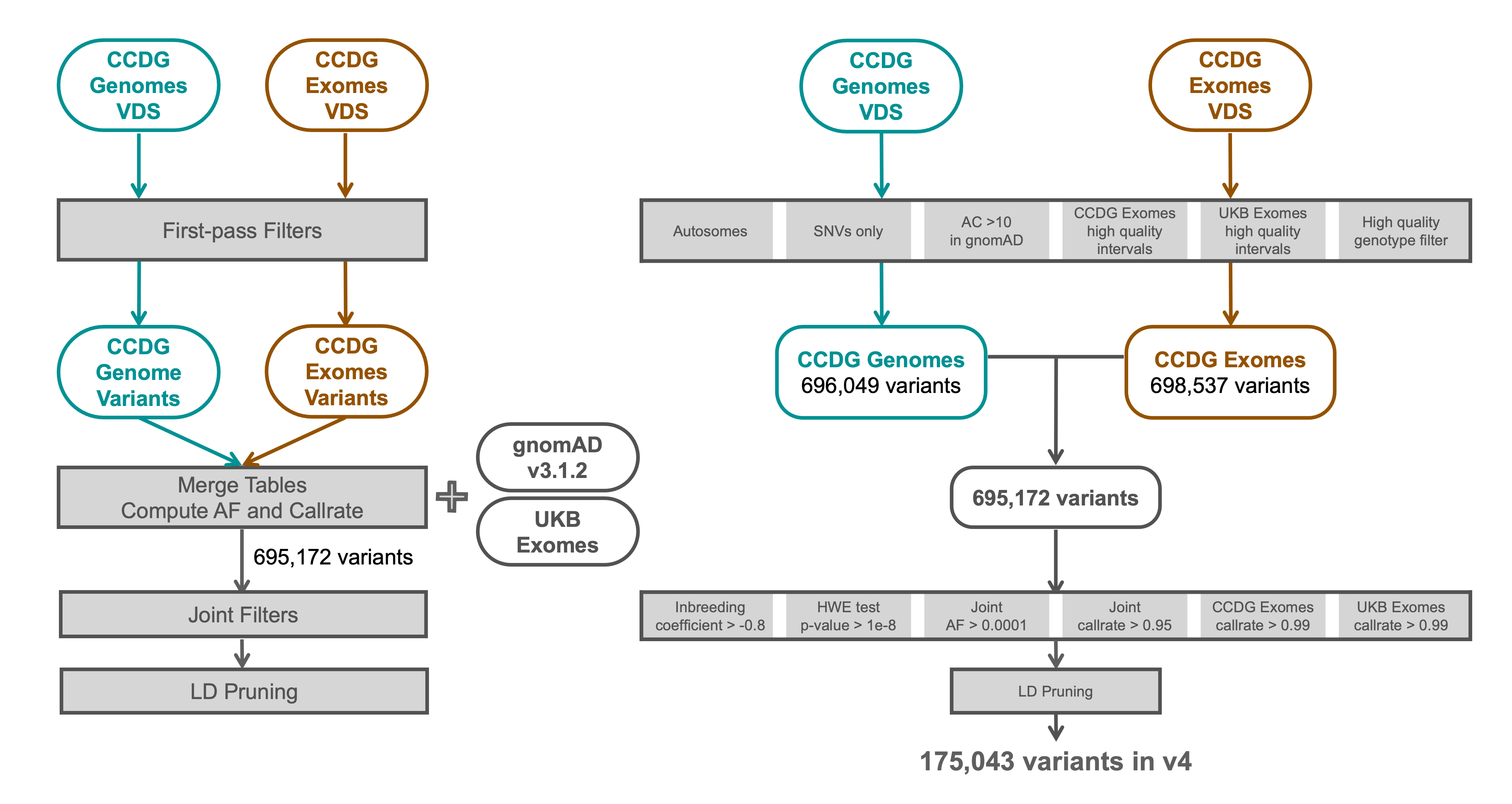
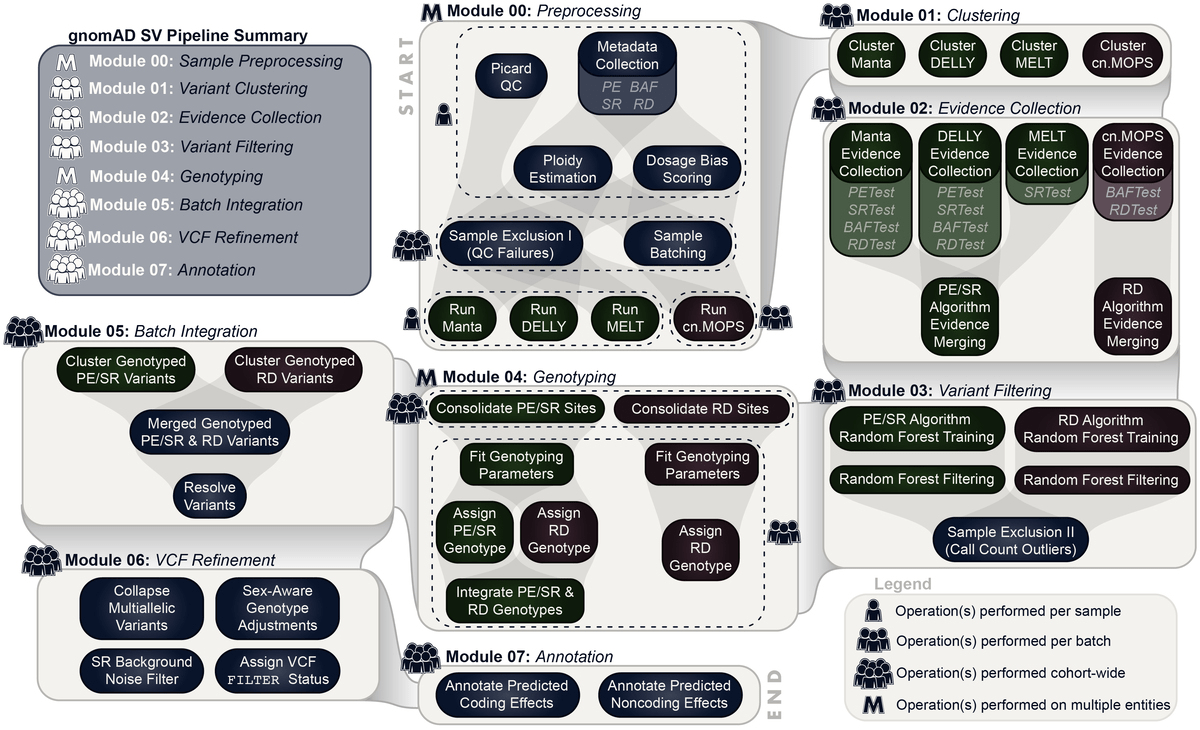
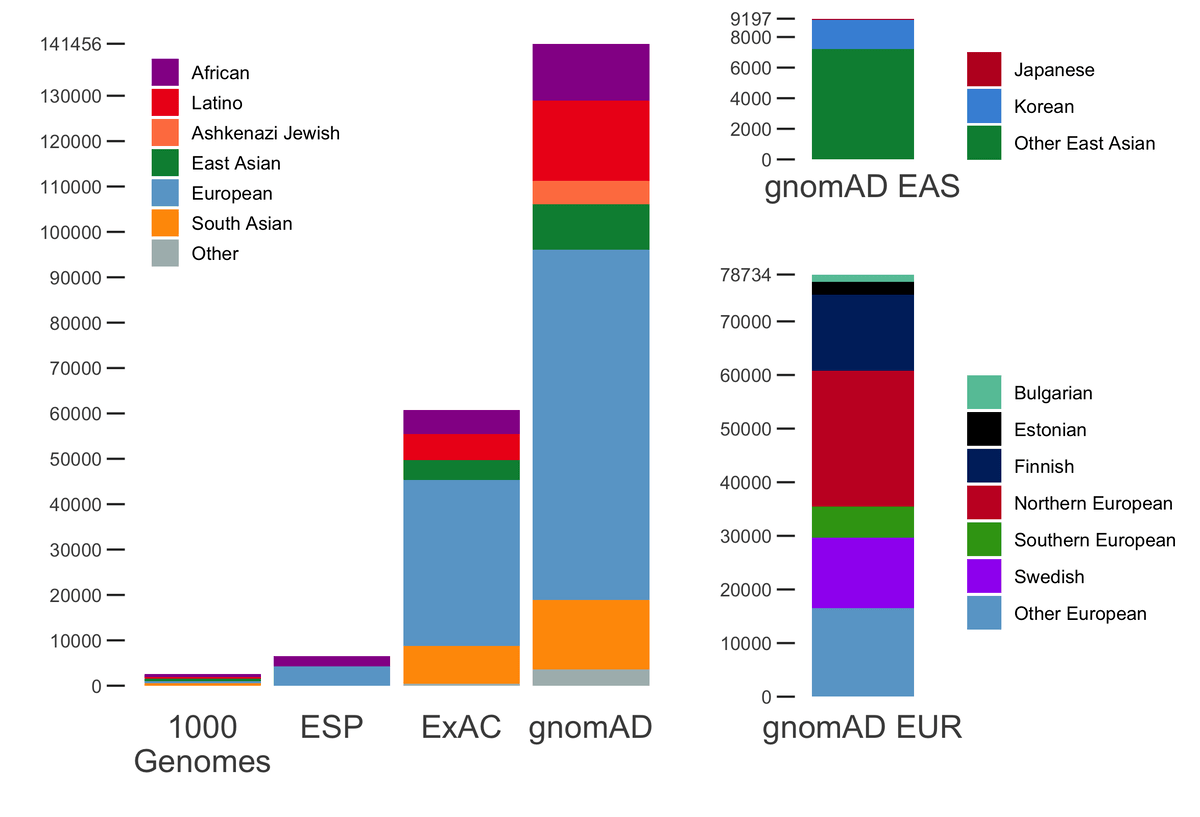
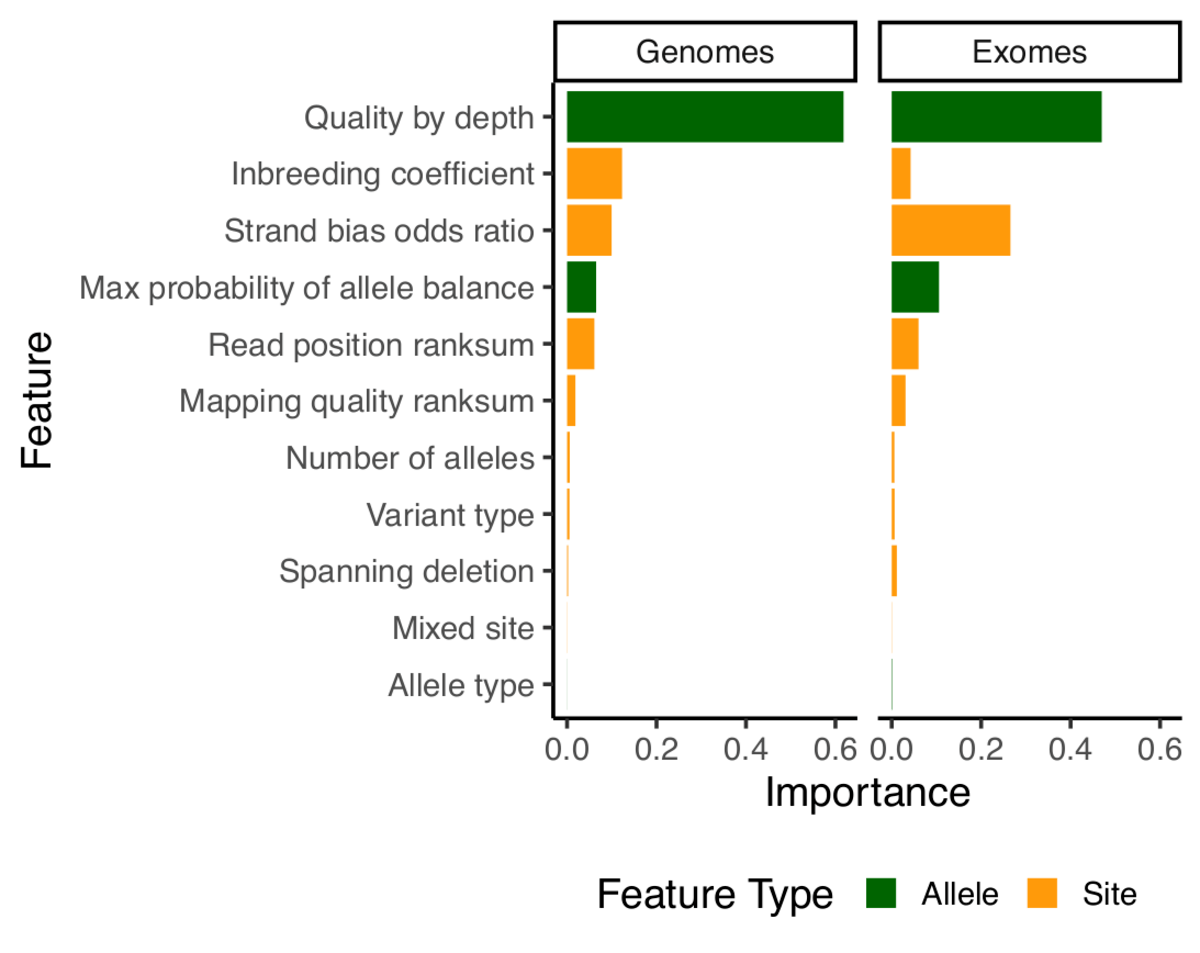
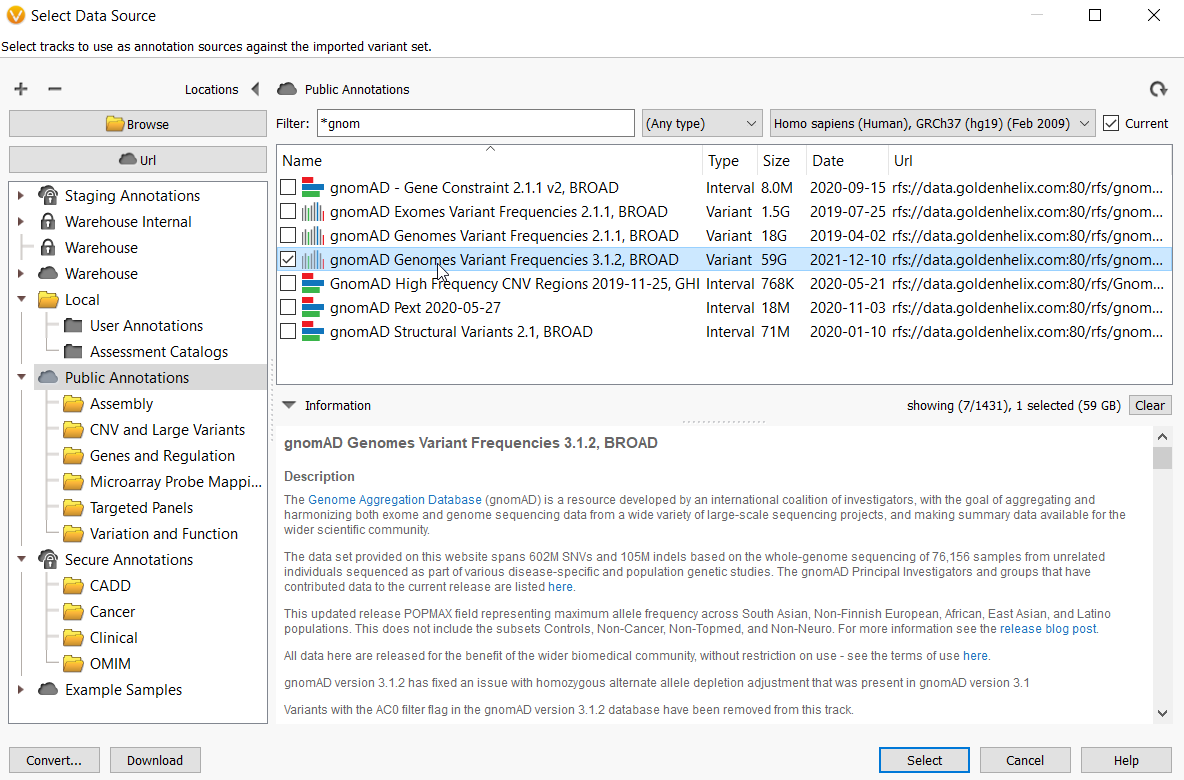
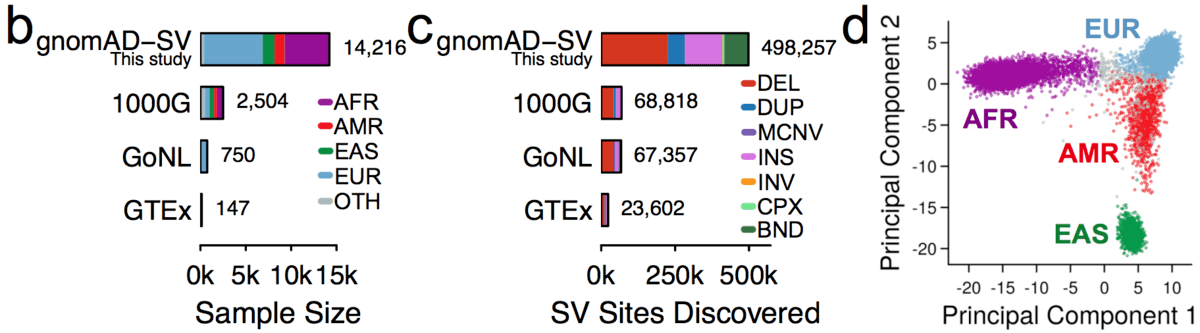
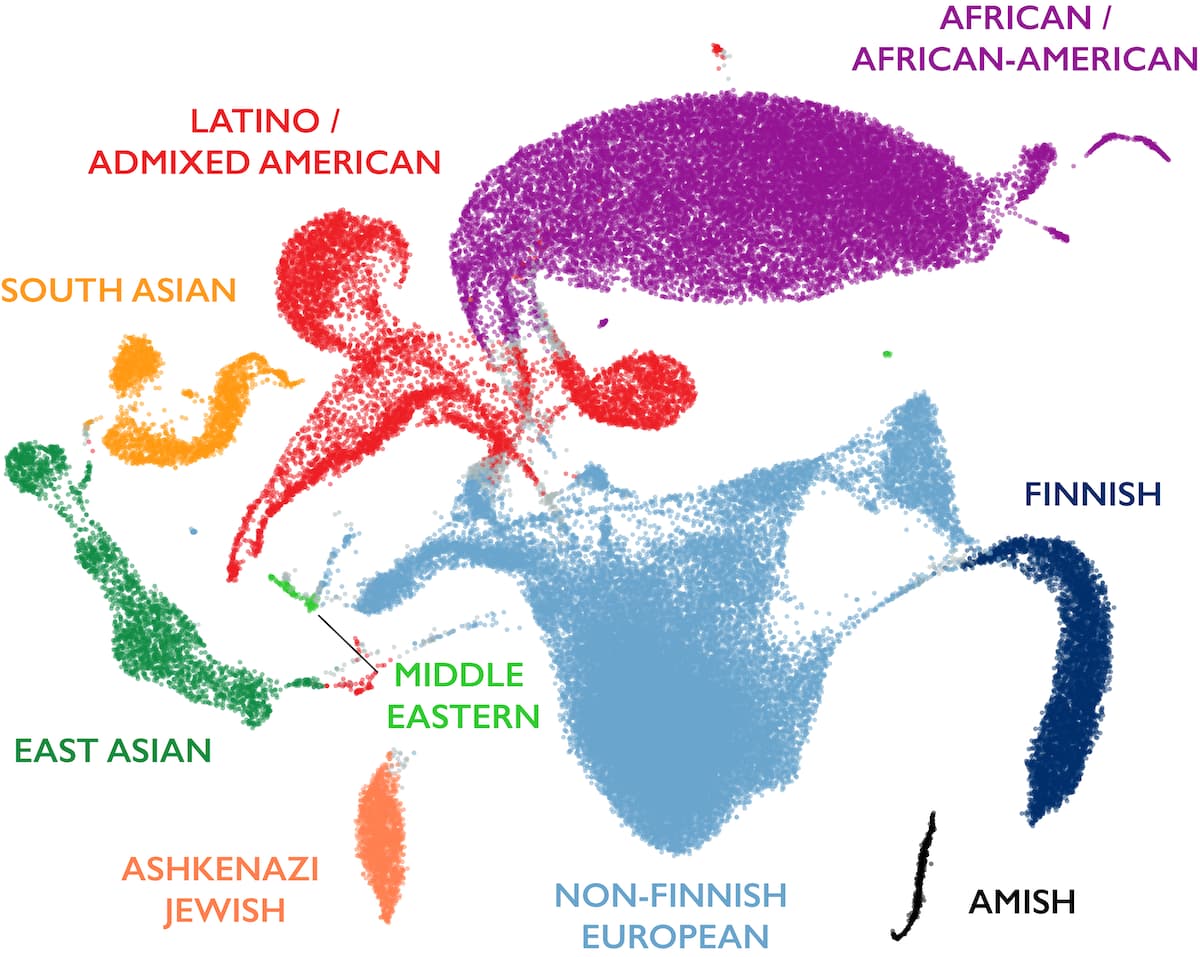
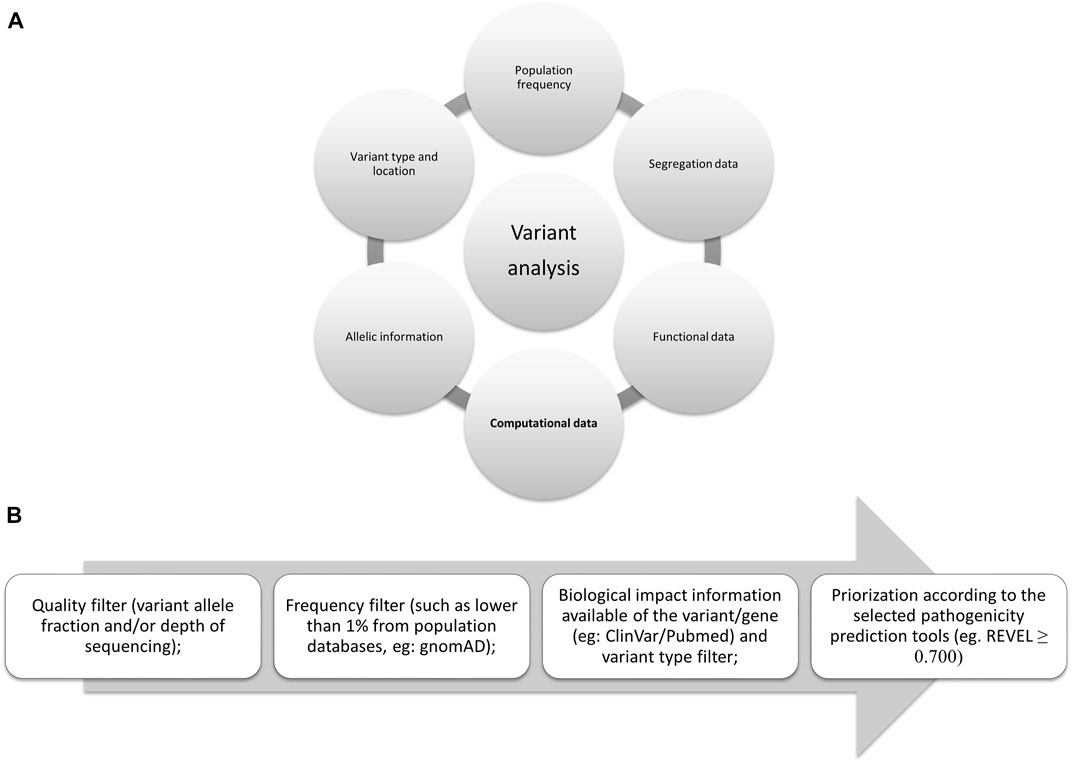
Variant interpretation using population databases: Lessons from gnomAD - Gudmundsson - 2022 - Human Mutation - Wiley Online Library

Systematic evaluation of gene variants linked to hearing loss based on allele frequency threshold and filtering allele frequency | Scientific Reports

Genome Aggregation Database on X: "Another #gnomAD browser update! On variant pages you can now quickly switch between genome builds and gnomAD versions using our new liftover feature. This is available on
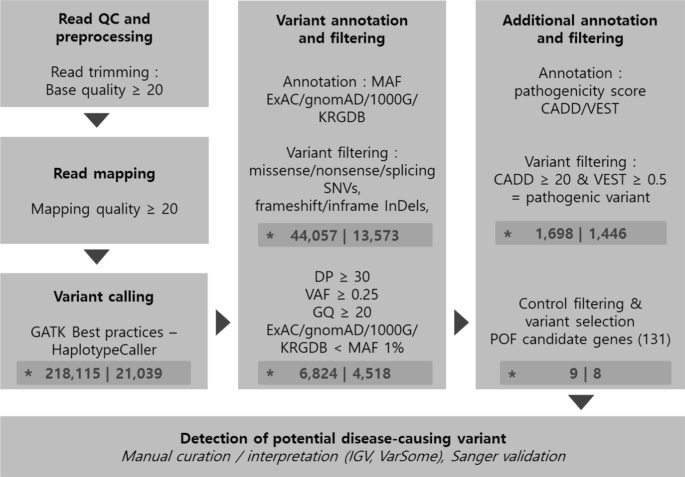
Identification of potential causal variants for premature ovarian failure by whole exome sequencing | BMC Medical Genomics | Full Text
Comparison of observed and gnomAD filtering allele frequencies (AFs)... | Download Scientific Diagram
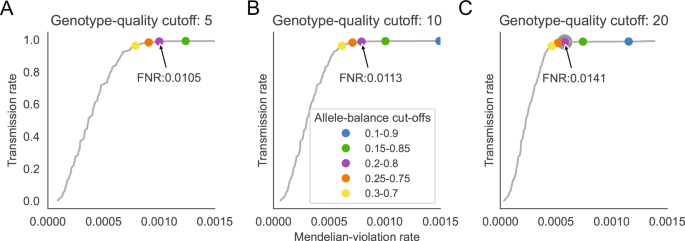
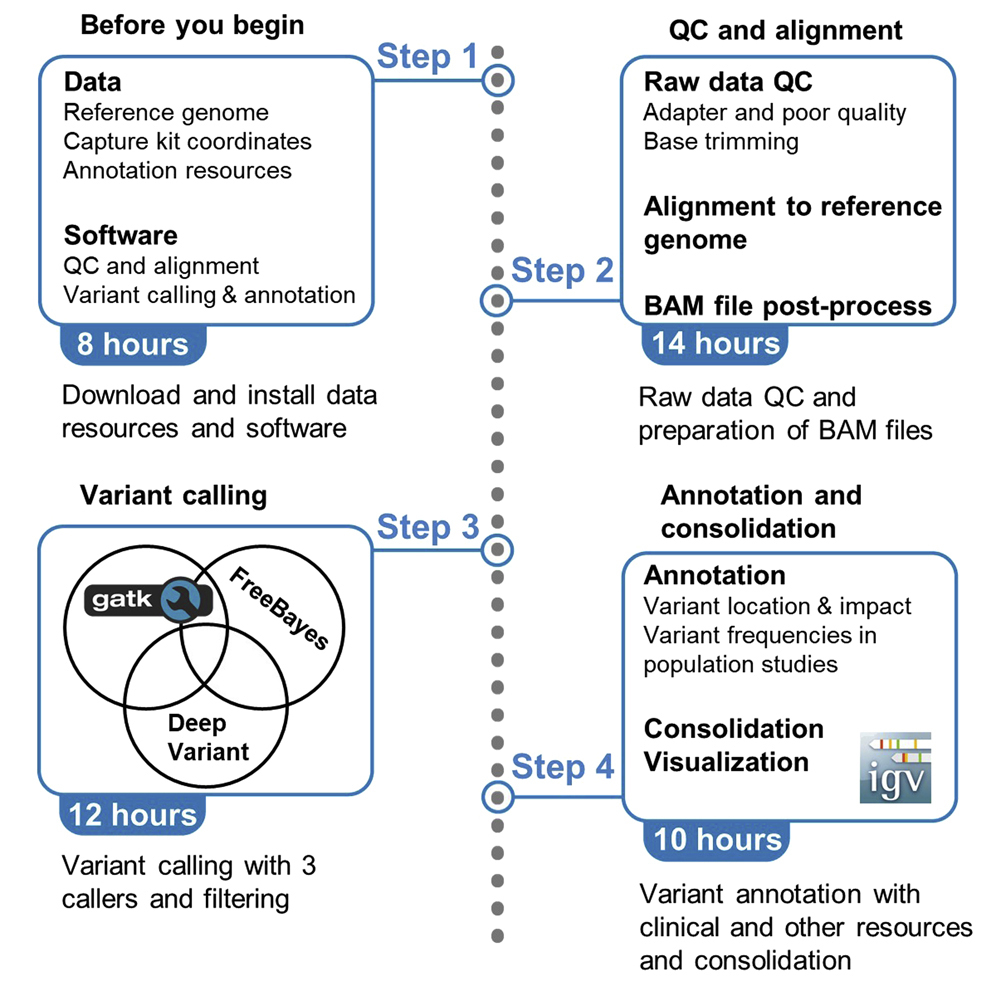
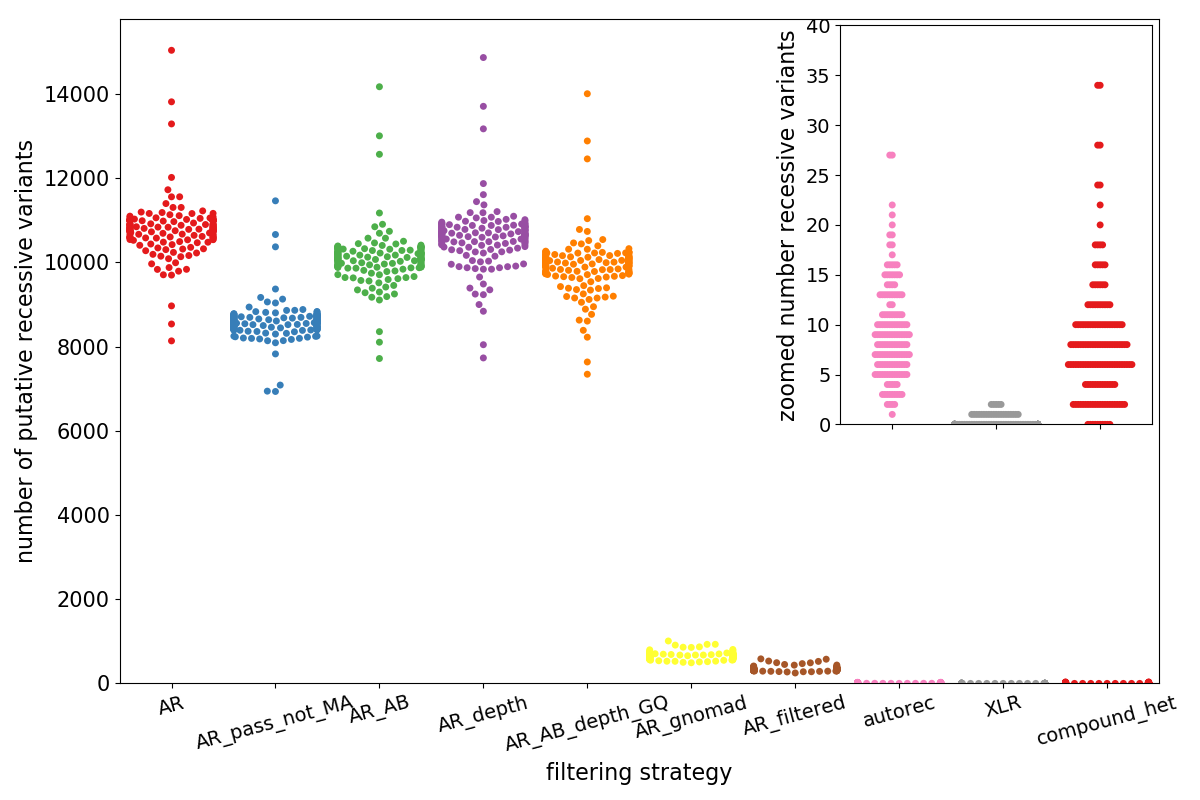
Effective variant filtering and expected candidate variant yield in studies of rare human disease | npj Genomic Medicine

Effective variant filtering and expected candidate variant yield in studies of rare human disease | bioRxiv

Variant interpretation using population databases: Lessons from gnomAD - Gudmundsson - 2022 - Human Mutation - Wiley Online Library

Variant filtering, digenic variants, and other challenges in clinical sequencing: a lesson from fibrillinopathies - Najafi - 2020 - Clinical Genetics - Wiley Online Library

Genome Aggregation Database on X: "We heard your feedback and updated the way #ClinVar data is displayed on #gnomAD browser. The ClinVar Track now includes: - Displaying variant type through shapes -
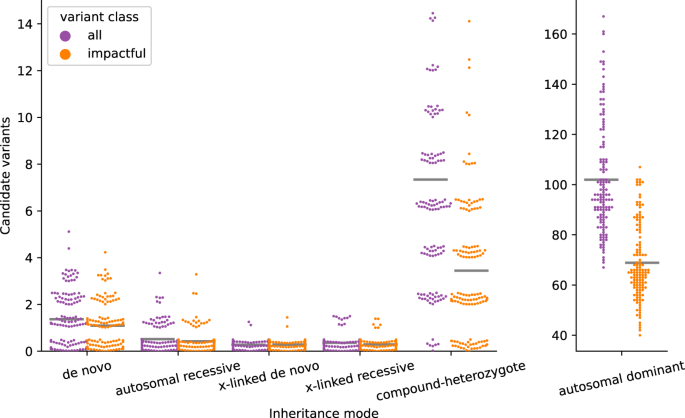
Effective variant filtering and expected candidate variant yield in studies of rare human disease | npj Genomic Medicine