A deconvolution method and its application in analyzing the cellular fractions in acute myeloid leukemia samples | BMC Genomics | Full Text

Bar chart representing the top 30 functionally enriched gene ontology... | Download Scientific Diagram

List of the 5951 SNP markers selected with their gene ontology (GO) for... | Download Scientific Diagram

Genetic variation in heat tolerance of the coral Platygyra daedalea indicates potential for adaptation to ocean warming | bioRxiv

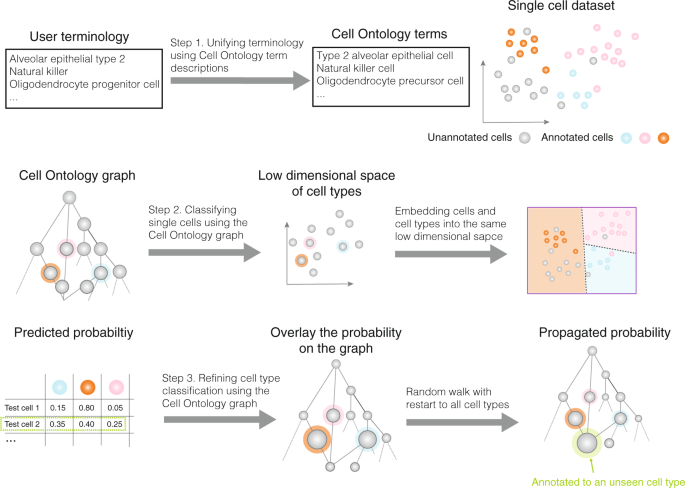
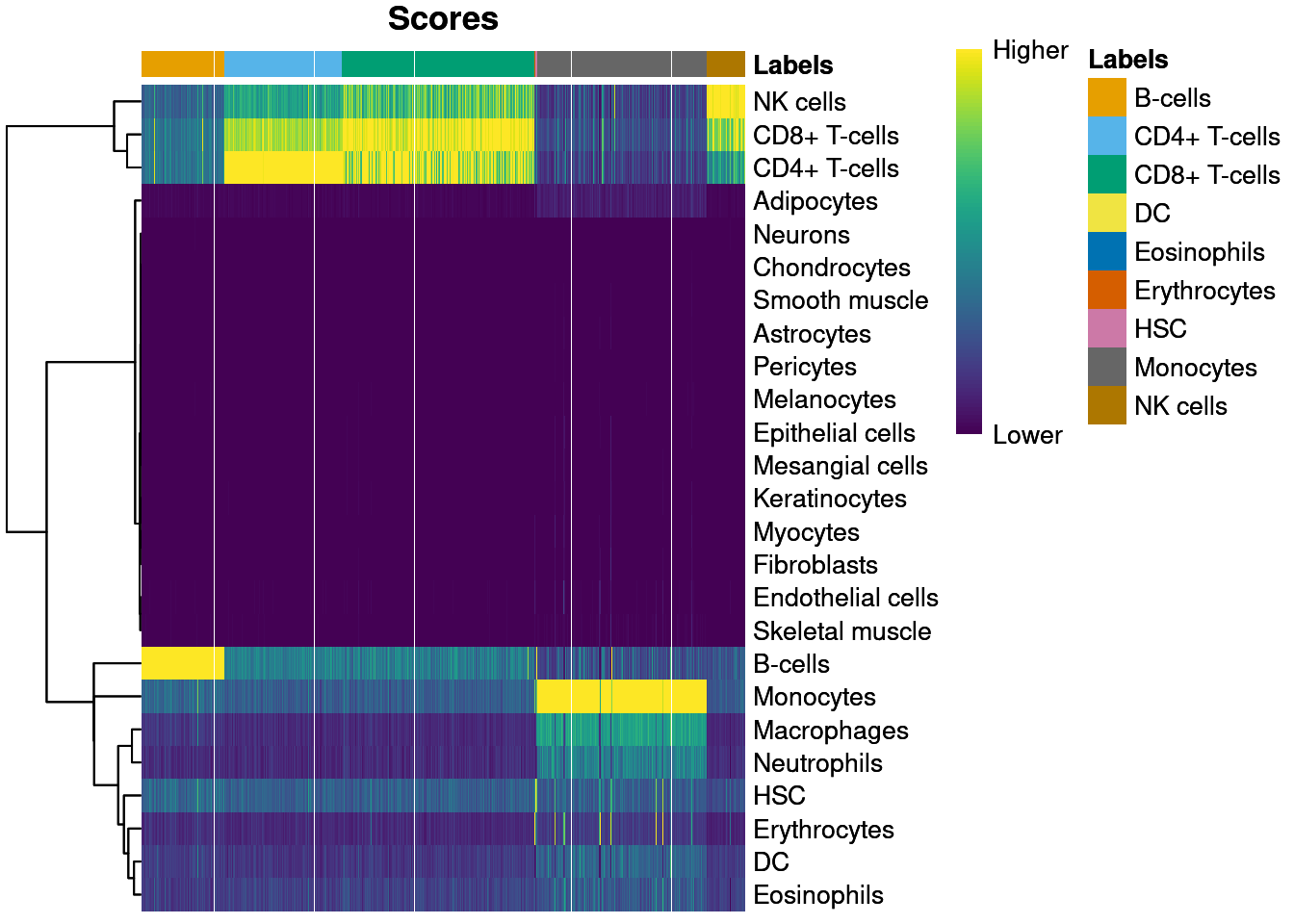
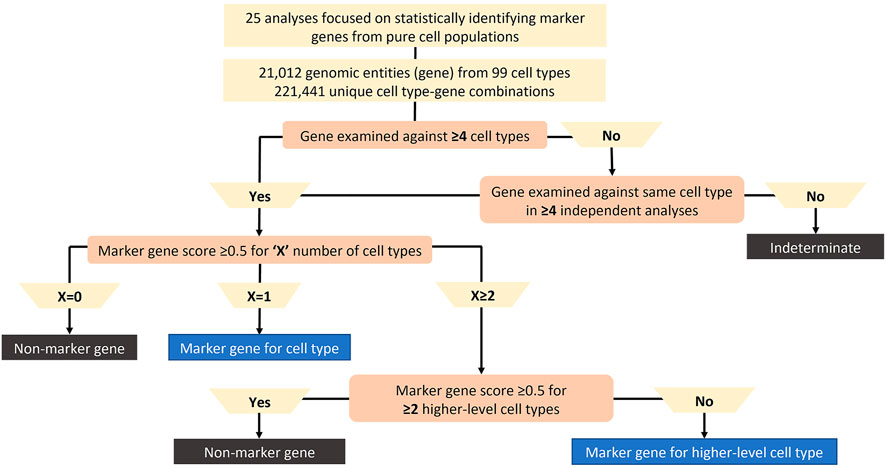
Automated methods for cell type annotation on scRNA-seq data - Computational and Structural Biotechnology Journal
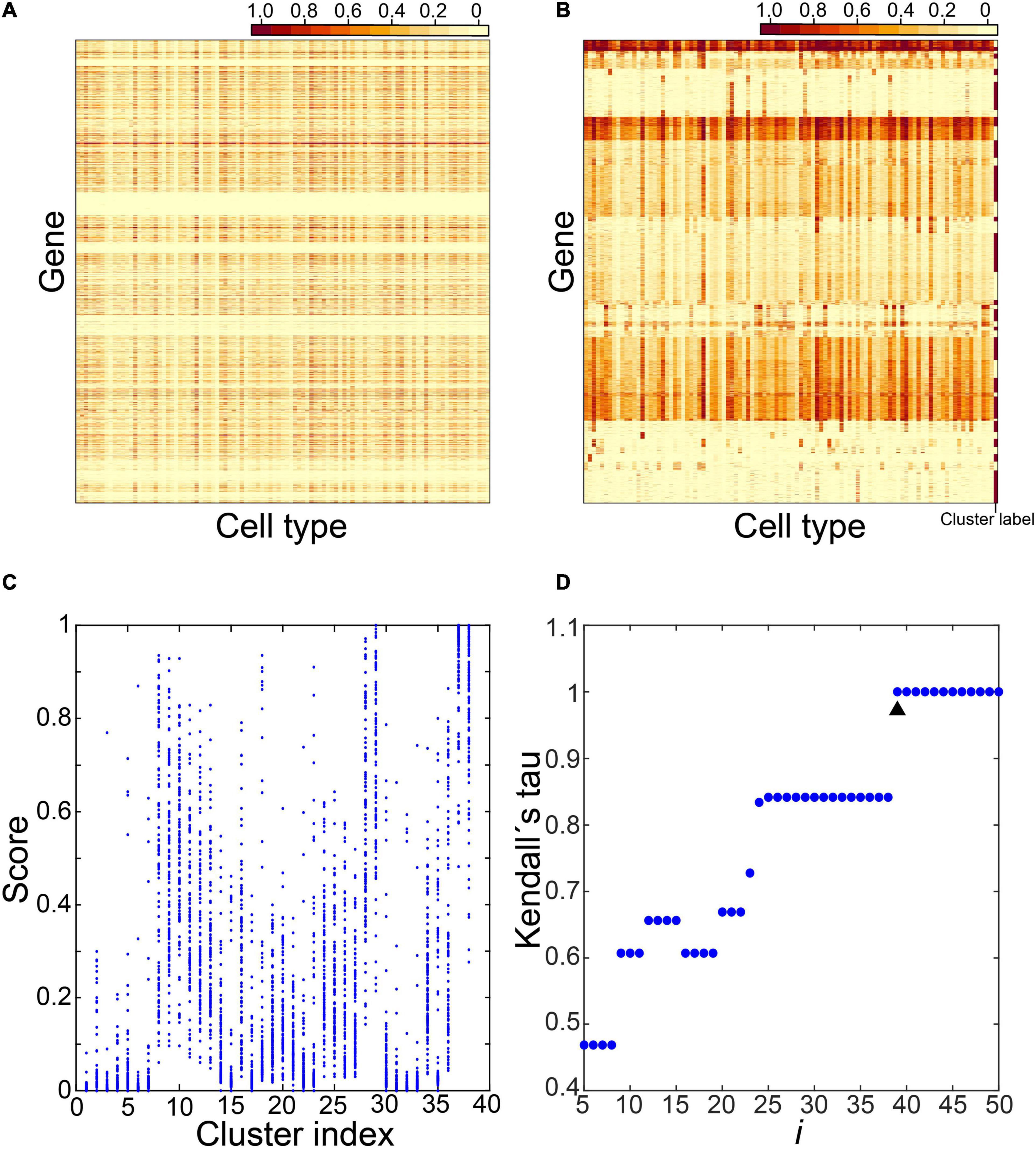
Frontiers | Using Cell Type–Specific Genes to Identify Cell-Type Transitions Between Different in vitro Culture Conditions

Distribution of gene ontology secondary markers for DEPs in the VD group. | Download Scientific Diagram

How to perform signaling pathway and Gene ontology analysis using R in scRNA-seq data? · satijalab seurat · Discussion #6080 · GitHub
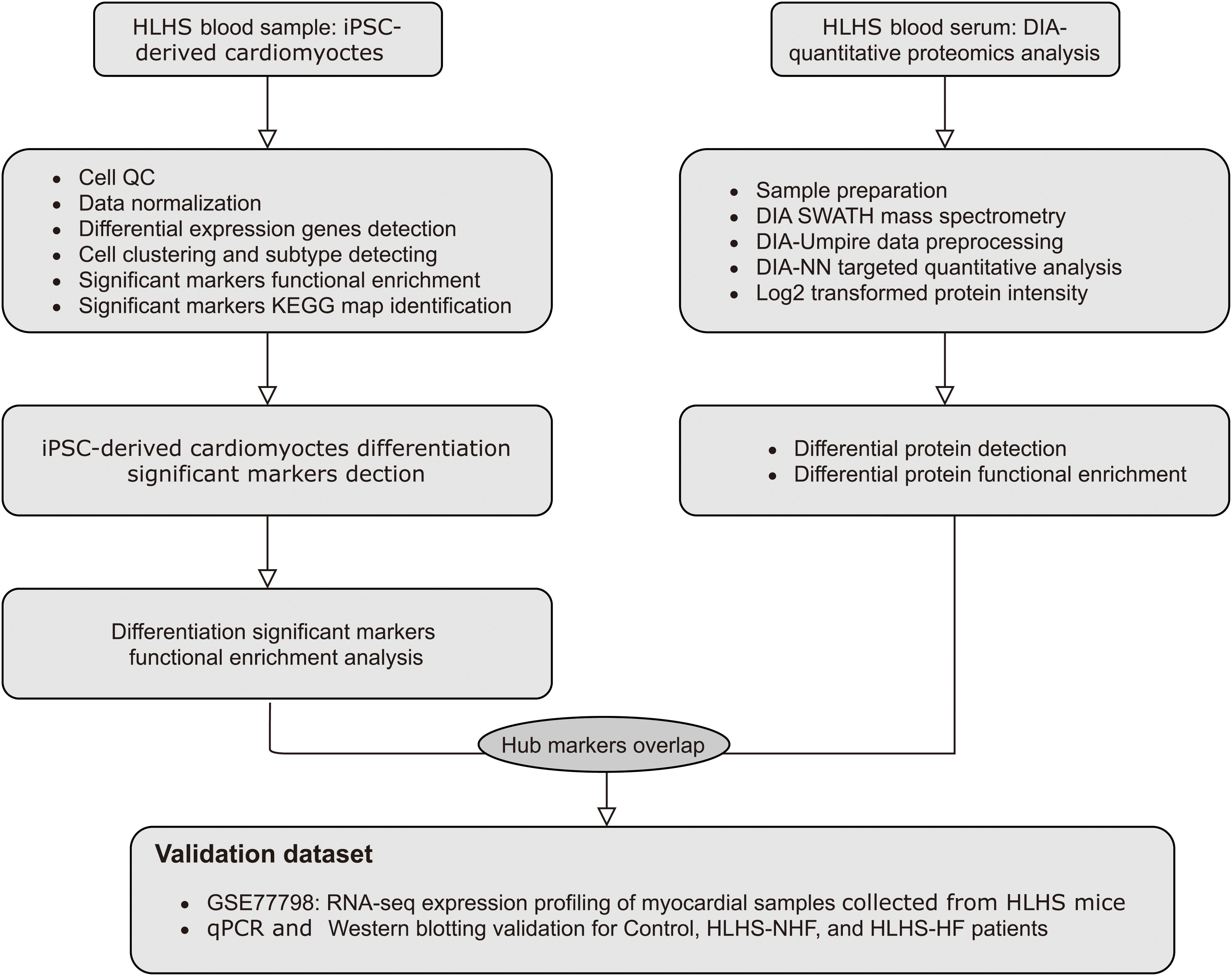
Frontiers | Single-Cell RNA Sequencing and Quantitative Proteomics Analysis Elucidate Marker Genes and Molecular Mechanisms in Hypoplastic Left Heart Patients With Heart Failure
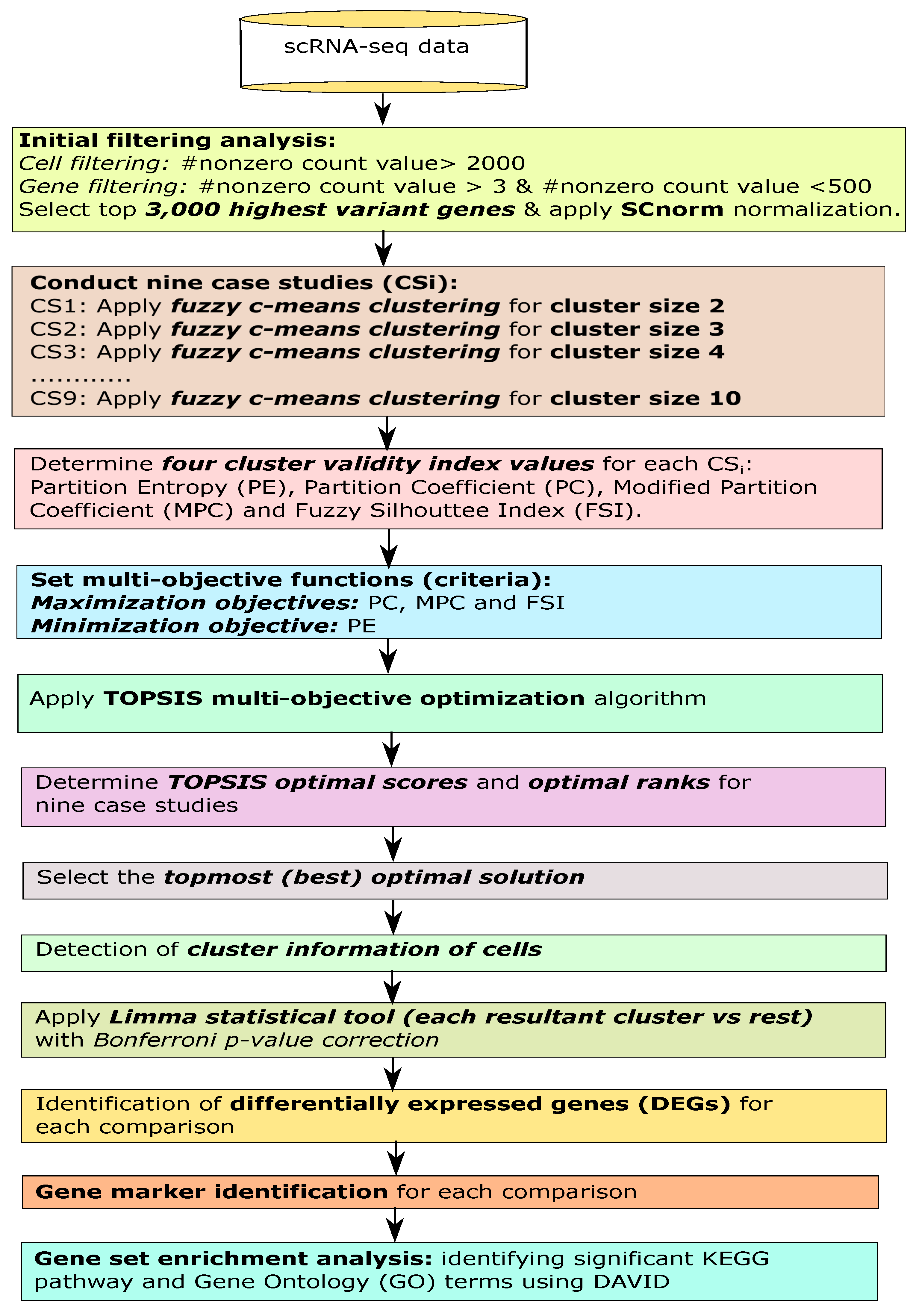
Genes | Free Full-Text | Multi-Objective Optimized Fuzzy Clustering for Detecting Cell Clusters from Single-Cell Expression Profiles

Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis | Gene Therapy

Identification of lung cancer gene markers through kernel maximum mean discrepancy and information entropy | BMC Medical Genomics | Full Text