Frontiers | Computational models for prediction of protein–protein interaction in rice and Magnaporthe grisea

Methods for Systematic Identification of Membrane Proteins for Specific Capture of Cancer-Derived Extracellular Vesicles - ScienceDirect

The graphical output of TMHMM, showing the posterior probabilities for... | Download Scientific Diagram
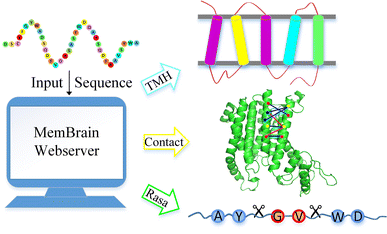
MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | Nano-Micro Letters

Topology Prediction Improvement of α-helical Transmembrane Proteins Through Helix-tail Modeling and Multiscale Deep Learning Fusion - ScienceDirect

Machine learning in computational modelling of membrane protein sequences and structures: From methodologies to applications - ScienceDirect
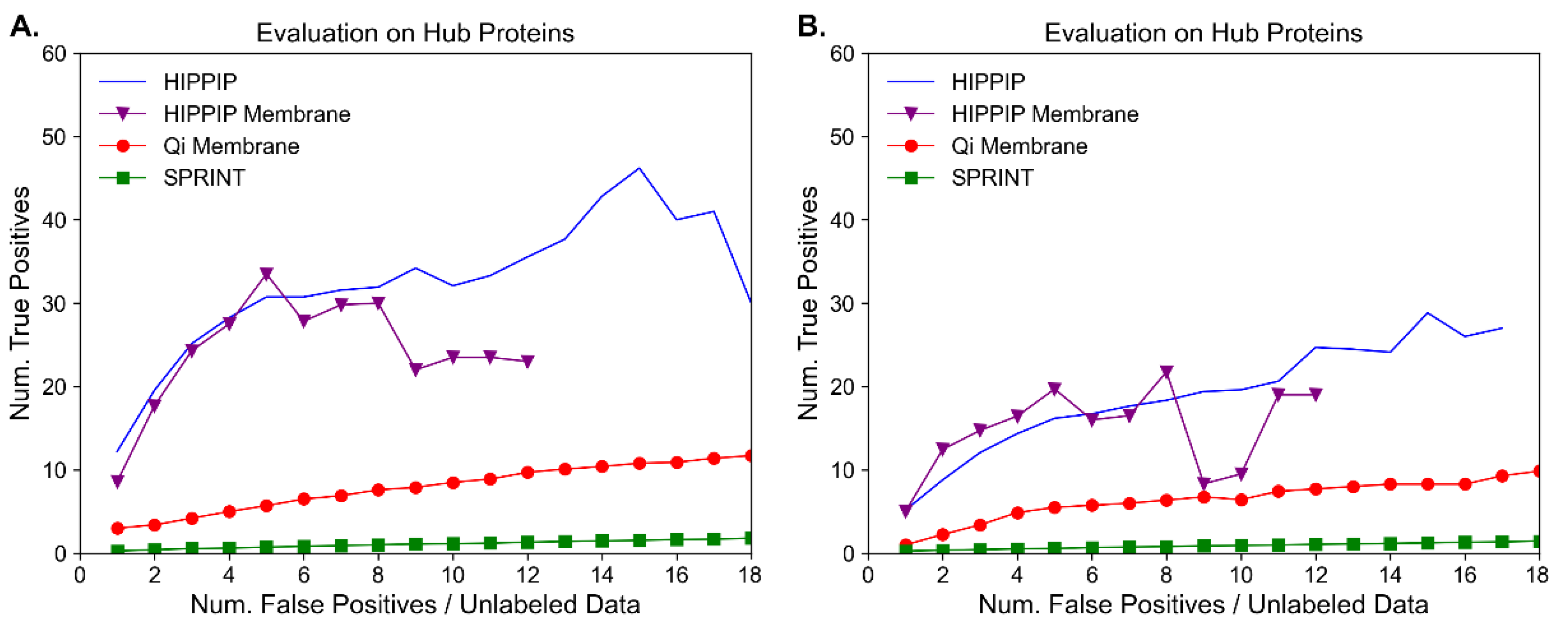
Molecules | Free Full-Text | Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms

MutTMPredictor: Robust and accurate cascade XGBoost classifier for prediction of mutations in transmembrane proteins - Computational and Structural Biotechnology Journal

MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | Nano-Micro Letters

Frontiers | Identifying well-folded de novo proteins in the new era of accurate structure prediction
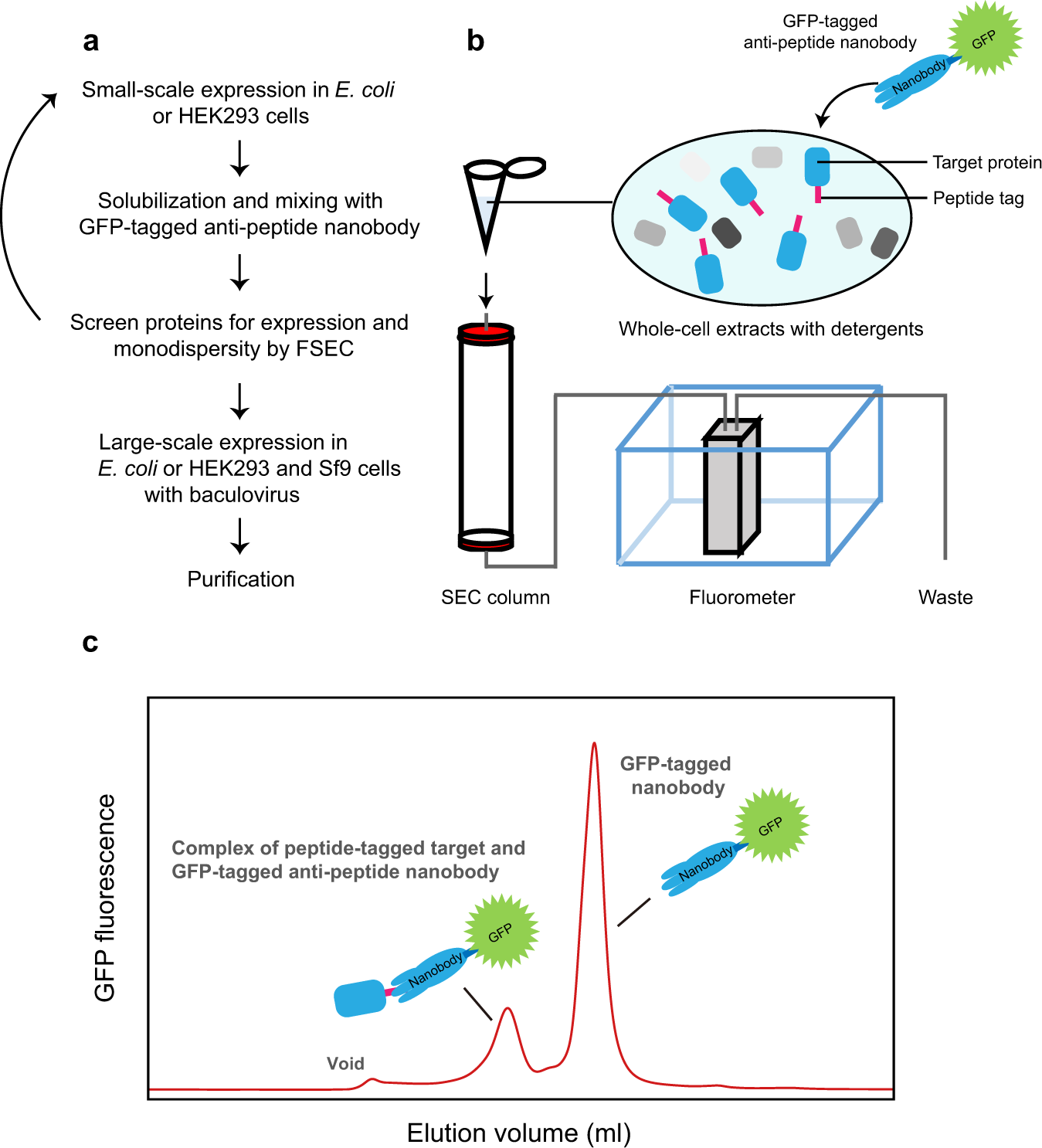
Fluorescence-detection size-exclusion chromatography utilizing nanobody technology for expression screening of membrane proteins | Communications Biology
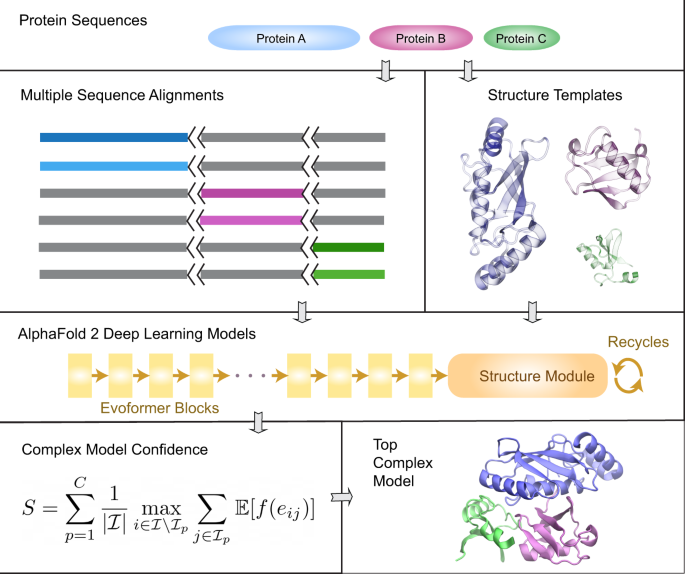
AF2Complex predicts direct physical interactions in multimeric proteins with deep learning | Nature Communications

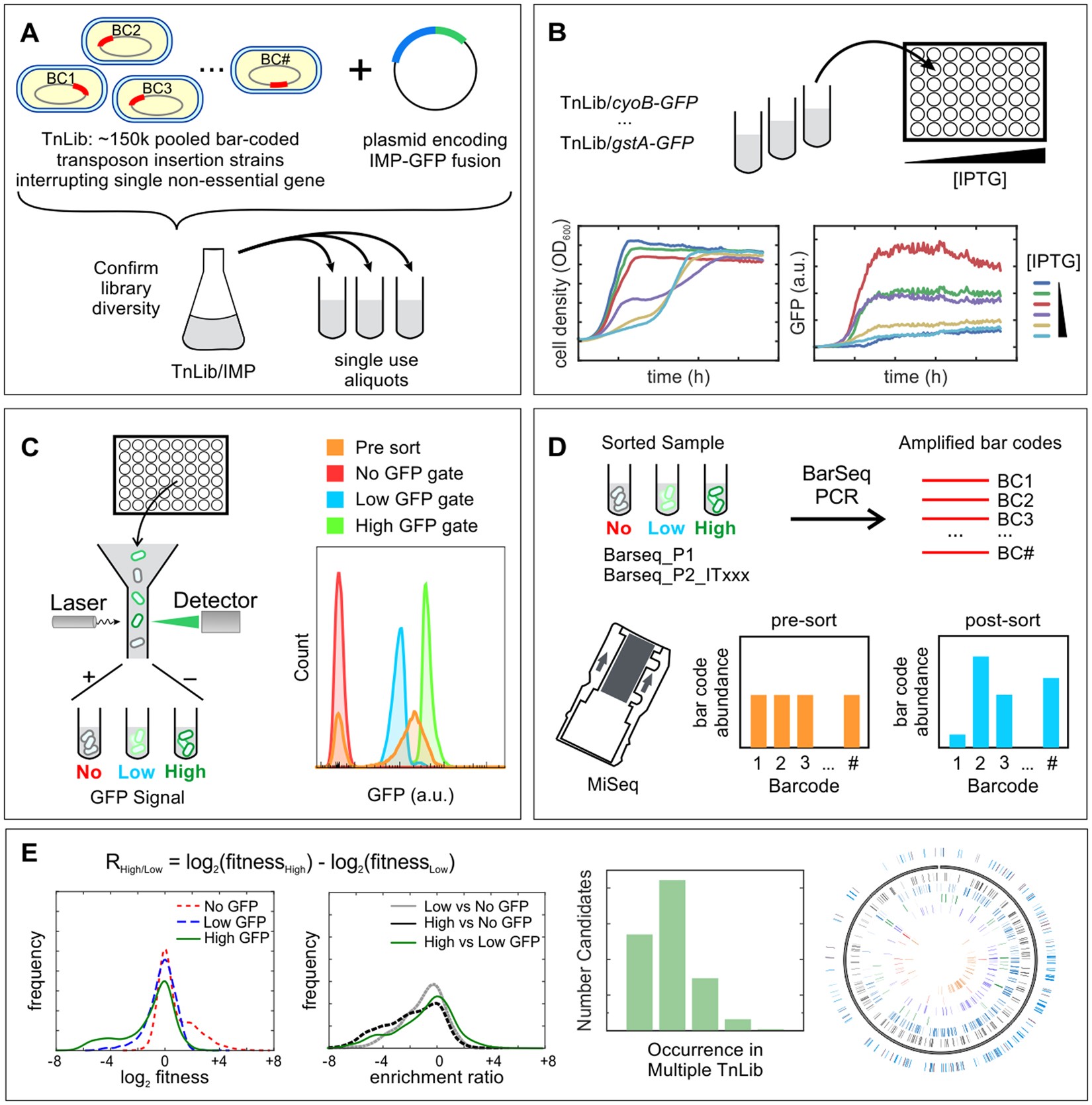
![RFPDR: a random forest approach for plant disease resistance protein prediction [PeerJ] RFPDR: a random forest approach for plant disease resistance protein prediction [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/11683/1/fig-2-full.png)